ALLOTYPIC VARIATION IN ANTIGEN PROCESSING CONTROLS ANTIGENIC PEPTIDE GENERATION FROM SARS-COV-2 S1 SPIKE GLYCOPROTEIN

Population genetic variability in immune system genes can often underlie variability in immune responses to pathogens. Cytotoxic T-lymphocytes are emerging as critical determinants of both SARS-CoV-2 infection severity and long-term immunity, after either recovery or vaccination. A hallmark of COVID-19 is its highly variable severity and breadth of immune responses between individuals. To address the underlying mechanisms behind this phenomenon we analyzed the proteolytic processing of S1 spike glycoprotein precursor antigenic peptides across 10 common allotypes of ER aminopeptidase 1 (ERAP1), a polymorphic intracellular enzyme that can regulate cytotoxic T-lymphocyte responses by generating or destroying antigenic peptides. We utilized a systematic proteomic approach that allows the concurrent analysis of hundreds of trimming reactions in parallel, thus better emulating antigen processing in the cell. While all ERAP1 allotypes were capable of producing optimal ligands for MHC class I molecules, including known SARS-CoV-2 epitopes, they presented significant differences in peptide sequences produced, suggesting allotype-dependent sequence biases. Allotype 10, previously suggested to be enzymatically deficient, was rather found to be functionally distinct from other allotypes. Our findings suggest that common ERAP1 allotypes can be a major source of heterogeneity in antigen processing and through this mechanism contribute to variable immune responses in COVID-19. (J Biol Chem. 2021).
CONFORMATIONAL DYNAMICS LINKED TO DOMAIN CLOSURE AND SUBSTRATE BINDING EXPLAIN THE ERAP1 ALLOSTERIC REGULATION MECHANISM

The endoplasmic-reticulum aminopeptidase ERAP1 processes antigenic peptides for loading on MHC-I proteins and recognition by CD8 T cells as they survey the body for infection and malignancy. Crystal structures have revealed ERAP1 in either open or closed conformations, but whether these occur in solution and are involved in catalysis is not clear. Here, we assess ERAP1 conformational states in solution in the presence of substrates, allosteric activators, and inhibitors by small-angle X-ray scattering. We also characterize changes in protein conformation by X-ray crystallography, and we localize alternate C-terminal binding sites by chemical crosslinking. Structural and enzymatic data suggest that the structural reconfigurations of ERAP1 active site are physically linked to domain closure and are promoted by binding of long peptide substrates. These results clarify steps required for ERAP1 catalysis, demonstrate the importance of conformational dynamics within the catalytic cycle, and provide a mechanism for the observed allosteric regulation and Lys/Arg528 polymorphism disease association. (Nature Communications 2021).
COMMON ALLOTYPES OF ER AMINOPEPTIDASE 1 HAVE SUBSTRATE-DEPENDENT AND HIGHLY VARIABLE ENZYMATIC PROPERTIES
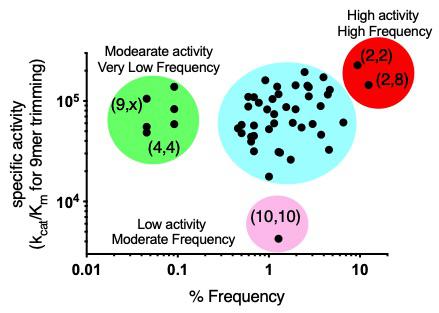
Polymorphic variation of immune system proteins can drive variability of individual immune responses. ER aminopeptidase 1 (ERAP1) generates antigenic peptides for presentation by MHC class I molecules. Coding single nucleotide polymorphisms (SNPs) in ERAP1 have been associated with predisposition to inflammatory rheumatic disease and shown to affect functional properties of the enzyme, but the interplay between combinations of these SNPs as they exist in allotypes, has not been thoroughly explored. We used phased genotype data to estimate ERAP1 allotype frequency in 2,504 individuals across five major human populations, generated highly pure recombinant enzymes corresponding to the 10 most common ERAP1 allotypes and systematically characterized their in vitro enzymatic properties. We find that ERAP1 allotypes possess a wide range of enzymatic activities, up to 60-fold, whose ranking is substrate-dependent. Strikingly, allotype 10, previously associated with Behcet's disease, is consistently a low-activity outlier, suggesting that a significant percentage of individuals carry a sub-active ERAP1 gene. Enzymatic analysis revealed that ERAP1 allotypes can differ in both catalytic efficiency and substrate affinity, differences that can change intermediate accumulation in multi-step trimming reactions. Alterations in efficacy of an allosteric inhibitor that targets the regulatory site suggest that allotypic variation influences the communication between the regulatory and the active site. Our work defines the wide landscape of ERAP1 activity in human populations and demonstrates how common allotypes can induce substrate-dependent variability in antigen processing, thus contributing, in synergy with MHC haplotypes, to immune response variability and to predisposition to chronic inflammatory conditions (JBC 2021).
GENERATION OF SARS-COV-2 S1 SPIKE GLYCOPROTEIN PUTATIVE ANTIGENIC EPITOPES IN VITRO BY INTRACELLULAR AMINOPEPTIDASES
Presentation of antigenic peptides by MHCI is central to cellular immune responses against viral pathogens. While adaptive immune responses versus SARS-CoV-2 can be of critical importance to both recovery and vaccine efficacy, how protein antigens from this pathogen are processed to generate antigenic peptides is largely unknown. Here, we analyzed the proteolytic processing of overlapping precursor peptides spanning the entire sequence of the S1 spike glycoprotein of SARS-CoV-2, by three key enzymes that generate antigenic peptides, aminopeptidases ERAP1, ERAP2 and IRAP. All enzymes generated shorter peptides with sequences suitable for binding onto HLA alleles, but with distinct specificity fingerprints. ERAP1 was the most efficient in generating peptides 8-11 residues long, the optimal length for HLA binding, while IRAP was the least efficient. The combination of ERAP1 with ERAP2 greatly limited the variability of peptide sequences produced. Less than 7% of computationally predicted epitopes were found to be produced experimentally, suggesting that aminopeptidase processing may constitute a significant filter to epitope presentation. These experimentally generated putative epitopes could be prioritized for SARS-CoV-2 immunogenicity studies and vaccine design. We furthermore propose that this in vitro trimming approach could constitute a general filtering method to enhance the prediction robustness for viral antigenic epitopes (J Proteome Res).

Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims antigenic peptide precursors to generate mature antigenic peptides for presentation by major histocompatibility complex class I (MHCI) molecules and regulates adaptive immune responses. ERAP1 has been proposed to trim peptide precursors both in solution and in pre-formed MHCI-peptide complexes, but which mode is more relevant to its biological function remains controversial. Here, we compared ERAP1-mediated trimming of antigenic peptide precursors in solution or when bound to three MHCI alleles, HLA-B*58, HLA-B*08 and HLA-A*02. For all MHCI-peptide combinations, peptide binding onto MHCI protected against ERAP1-mediated trimming. In only a single MHCI-peptide combination, trimming of an HLA-B*08-bound 12mer progressed at a considerable rate, albeit still slower than in solution. Results from thermodynamic, kinetic and computational analyses suggested that this 12mer is highly labile and that apparent on-MHC trimming rates are always slower than that of MHCI-peptide dissociation. Both ERAP2 and leucine aminopeptidase, an enzyme unrelated to antigen processing, could trim this labile peptide from pre-formed MHCI complexes as efficiently as ERAP1. A pseudopeptide analogue with high affinity for both HLA-B*08 and the ERAP1 active site could not promote the formation of a ternary ERAP1-MHCI-peptide complex. Similarly, no interactions between ERAP1 and purified peptide loading complex (PLC) were detected in the absence or presence of a pseudopeptide trap. We conclude that MHCI binding protects peptides from ERAP1 degradation and that trimming in solution, along with the dynamic nature of peptide binding to MHCI, are sufficient to explain ERAP1 processing of antigenic peptide precursors. (JBC 2020)

