To achieve this aim we use the following physical chemical methodologies:
a. Solid State Nuclear Magnetic Resonance in order to study the dynamic changes it causes the drug in the lipid bilayers. In addition, the orientation of drugs in lipid bilayers is sought using deuterium NMR in collaboration with synthetic chemists who deuterate specific sites of drugs.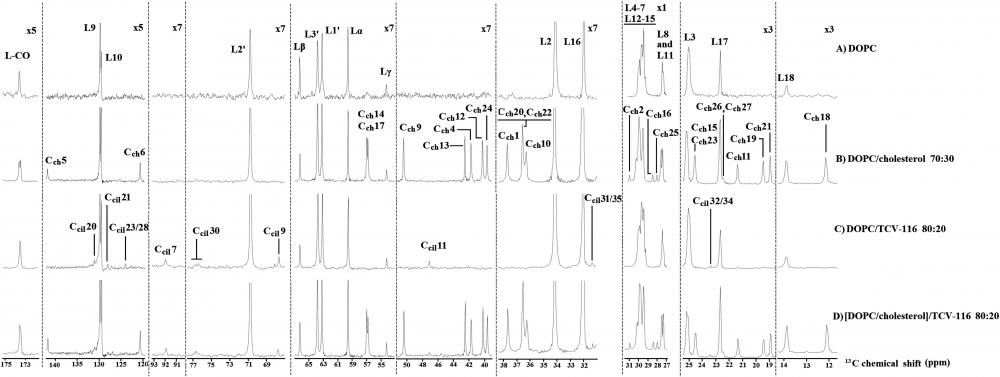
The application of solid-state NMR spectroscopy to study candesartan cilexetil (TCV-116) membrane interactions. Expanded regions of the 13C CP/MAS NMR spectra of (A) DOPC, (B) DOPC/cholesterol (molar ratio 70:30), (C) DOPC/TCV-116 (molar ratio 80:20) (D) [DOPC/cholesterol]/TCV-116 (molar ratio 80:20) obtained at 27 °C using 850 MHz Bruker spectrometer.
b. High Resolution Nuclear Magnetic Resonance in deuterated solvents or environments that simulate the active site of action.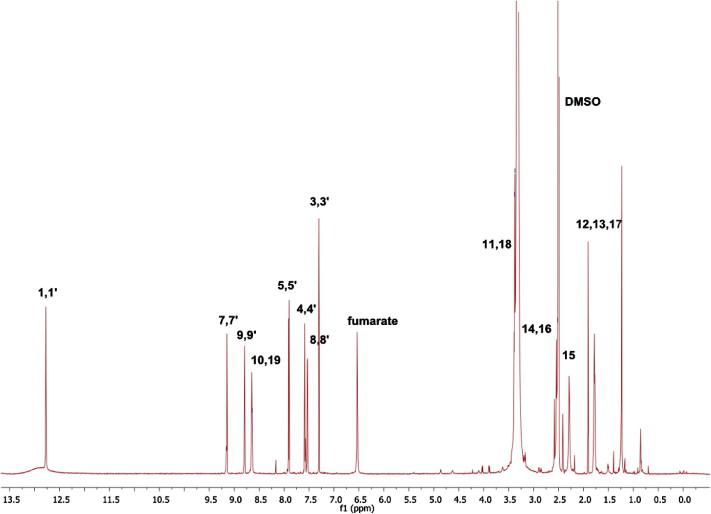
Analysis of two novel cytotoxic C2-substituted pyrrolo[2,3-f]quinolines in aqueous media, organic solvents, membrane bilayers and at the putative active site. 1H NMR spectrum of one the compounds obtained in DMSO-d6 at 25 °C and 800 MHz and the resulting conformations.
c. Conformational Analysis Using NMR ROE data in combination with computational analysis and molecular modeling to complement the experimental results.
The dynamic properties of angiotensin II type 1 receptor inverse agonists in solution and in the receptor site. Olmesartan's and olmesarten's ethyl ether conformations using NOE derived constraints.
d. Differential Scanning Calorimetry to study the thermal changes that causes the presence of pharmaceutical compounds with model membranes.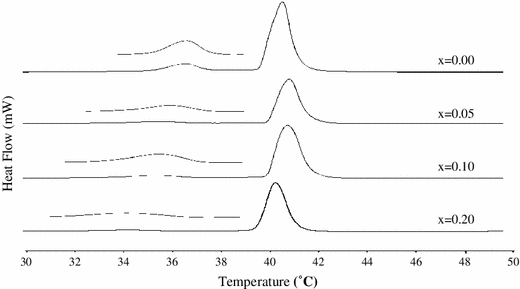
e. X-ray diffraction where the topography of drugs is sought in lipid bilayers. The X-ray diffraction experiments are run in Graz of Austria and ELETTRA facilities in Trieste of Italy.
f. Raman Spectroscopy to study the interactions of bioactive compounds with lipid bilayers. In particular, the trans:gauche ratio is estimated at the different mesomorphic states of lipid bilayers.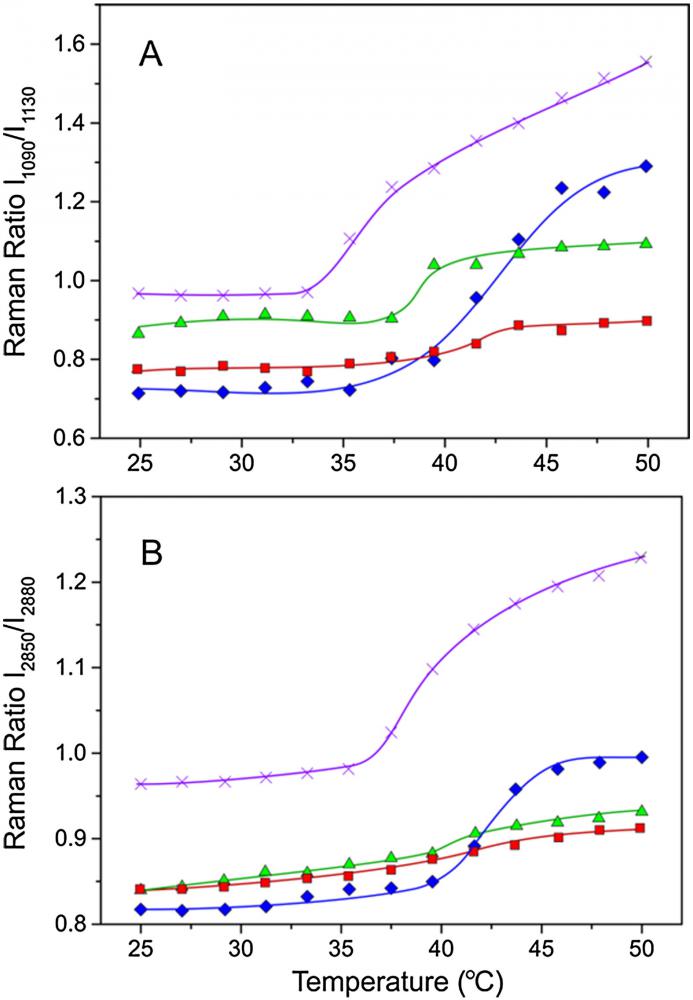
Comparative study of interactions of aliskiren and AT1 receptor antagonists with lipid bilayers. Raman Ratio I1090/I1130 vs. temperature plots for pure DPPC (![]() ), DPPC containing 20 mol.% of aliskiren (
), DPPC containing 20 mol.% of aliskiren (![]() ), DPPC containing 15 mol.% cholesterol (
), DPPC containing 15 mol.% cholesterol (![]() ) and [DPPC/cholesterol (85/15)]/aliskiren (80/20) (
) and [DPPC/cholesterol (85/15)]/aliskiren (80/20) (![]() ). The same symbolic is used for panel B representing I2850/I2880 vs. temperature plots.
). The same symbolic is used for panel B representing I2850/I2880 vs. temperature plots.
g. 3D-QSAR studies. CoMFA and CoMSIA softwares are applied to reveal the stereoelectronic requirements for a drug to exert biological activity.
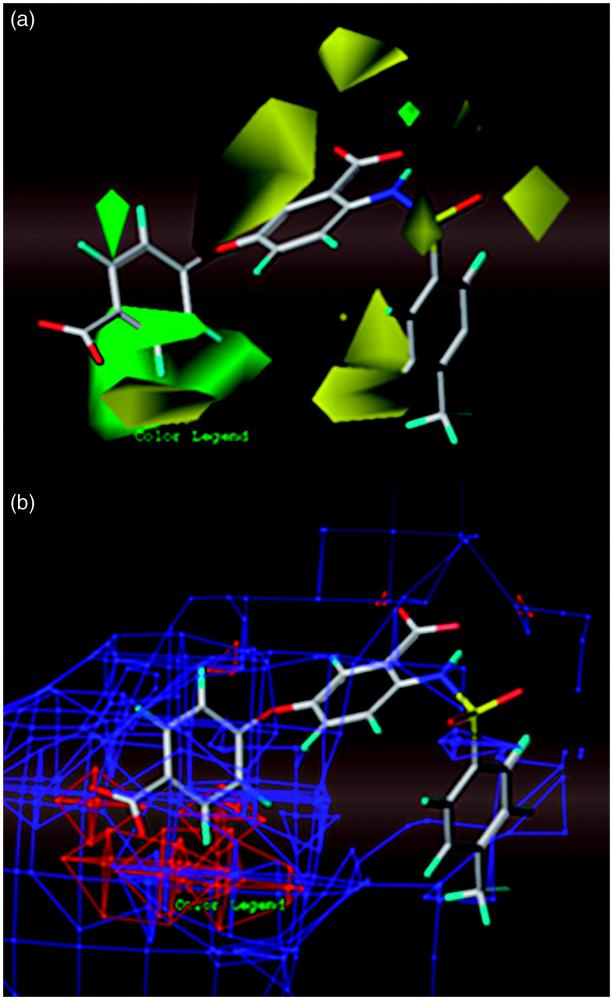
Searching for anthranilic acid-based thumb pocket 2 HCV NS5B polymerase inhibitors through a combination of molecular docking, 3D-QSAR and virtual screening. CoMFA StDev*Coeff contour maps around the active compound 8: (a) solid for steric field (green: bulky groups are favored; yellow: bulky groups are disfavored); (b) mesh for electrostatic field (red: electronegative groups are favored; blue: electropositive groups are favored).
h. Simulation of NMR spectra in liquid and solid state with the collaboration of theoretical physical chemists.
i. Docking studies of drugs acting on receptors, DNA and bilayer environment.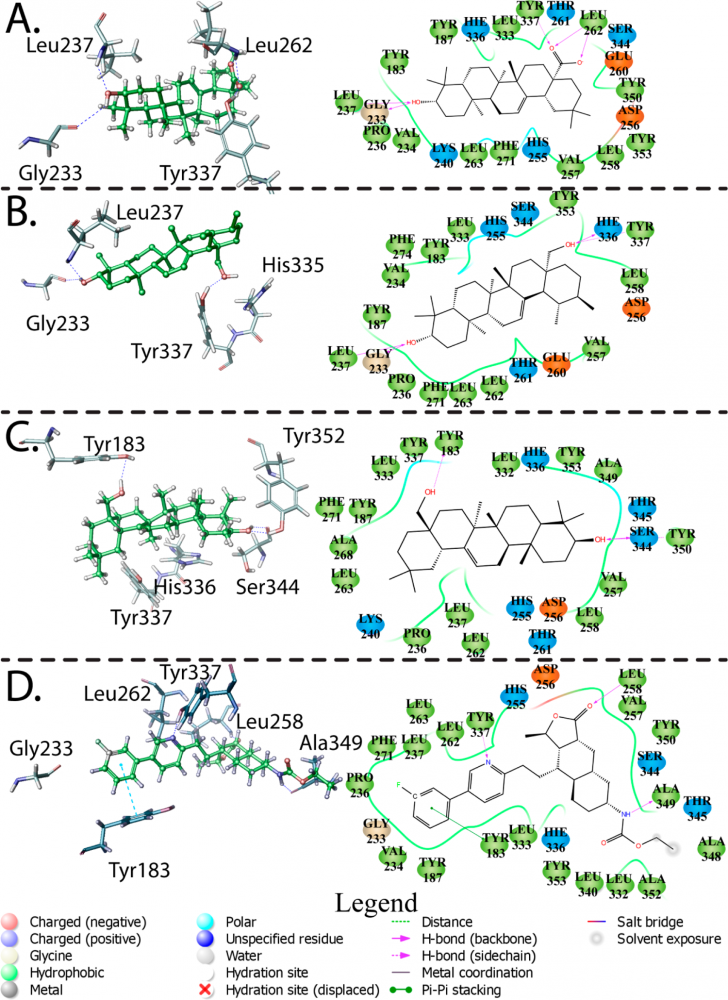
Deconvoluting the Dual Antiplatelet Activity of a Plant Extract. Predicted binding poses with best induced fit score of (A) oleanolic acid, (B) uvaol, (C) erythrodiol, and (D) the crystal structure of vorapaxar in PAR1.
Collaborators
Alexandros Makriyannis, Prof. Dr., (College of Science, Northeastern University)
Heinz Amenitsch, Ass.Prof. Dipl.-Ing. Dr.techn. (Institut für Anorganische Chemie, Elettra-Sincrotrone Trieste)
Simona Golic Grdadolnik, Prof. Dr., (Laboratory of Biomolecular Structure, National Institute of Chemistry, Slovenia)
Gregor Mali, Prof. Dr., (National Institute of Chemistry, Slovenia)
Michael Rappolt, Prof. Dr., (School of Food Science and Nutrition, University of Leeds)
Antreas Afantitis, Dr., (Novamechanics)
Andrew Tsotinis, Prof. Dr., (Faculty of Pharmacy, National and Kapodistrian University of Athens)
Ioannis Papanastasiou, Assistant Professor, (Faculty of Pharmacy, National and Kapodistrian University of Athens)

