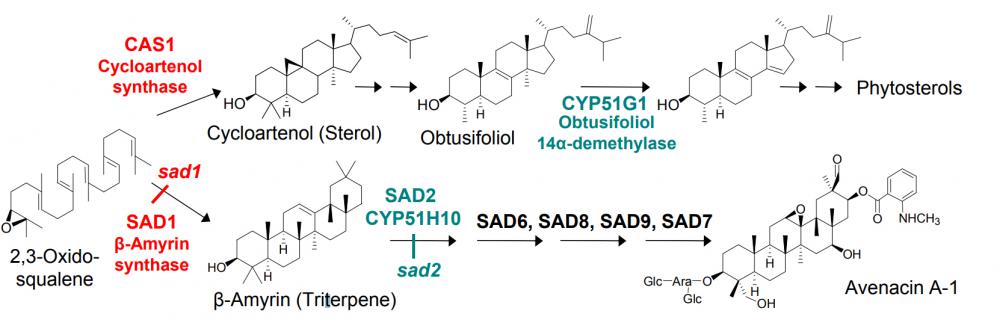
Many different plant species synthesise triterpenoid saponins as part of their normal programme of growth and development. Examples include plants that are exploited as sources of drugs, such as liquorice and ginseng, and also crop plants such as legumes and oats. Interest in these molecules stems from their medicinal properties, antimicrobial activity, and their likely role as determinants of plant disease resistance. Triterpenoid saponins are synthesised via the isoprenoid pathway by cyclization of 2,3-oxidosqualene to give primarily oleanane (β-amyrin) or dammarane triterpenoid skeletons. The triterpenoid backbone then undergoes various modifications (oxidation, substitution and glycosylation), mediated by cytochrome P450-dependent monooxygenases, glycosyltransferases and other enzymes. In general very little is known about the enzymes and biochemical pathways involved in saponin biosynthesis. The genetic machinery required for the elaboration of this important family of plant secondary metabolites is as yet largely uncharacterised, despite the considerable commercial interest in this important group of natural products. This is likely to be due in part to the complexity of the molecules and the lack of pathway intermediates for biochemical studies. Considerable advances have recently been made, however, in the area of 2,3-oxidosqualene cyclisation, and a number of genes encoding the enzymes that give rise to the diverse array of plant triterpenoid skeletons have been cloned. Progress has also been made in the characterisation of saponin glucosyltransferases.